People in the Cooley Lab
 Arielle Cooley. I am a broadly trained evolutionary biologist, primarily interested in the evolution of phenotypic diversity. My goal is to connect cellular, molecular and genomic processes to organismal variation in developmental phenotypes and fitness.
Arielle Cooley. I am a broadly trained evolutionary biologist, primarily interested in the evolution of phenotypic diversity. My goal is to connect cellular, molecular and genomic processes to organismal variation in developmental phenotypes and fitness.
My research background includes a post doc in Patricia Wittkopp's lab, where I focused on two closely related fruit fly species that differ in body color. My ongoing research interest, initiated as a graduate student in John Willis' lab, involves floral diversification in a group of South American wildflowers in the "monkeyflower" genus Mimulus. I am pursuing this work at Whitman College as well as teaching Genetics; Evolution & Development; Plant Physiology; and an Evolution course for nonmajors.
Lab alums
 Iker Sanchez Marquez '26 and Caine Ryan '26. Iker (left) and Caine (right) worked together to replicate an experimental comparison of gene function, between two alleles of the anthocyanin-activating transcription factor gene MYB5a. They quantified anthocyanin by both digital image analysis and spectrophotometry. Both approaches confirmed the results of our initial experiment: Coding sequence from the yellow-flowered M. l. luteus, suprisingly, drives slightly but significantly more anthocyanin production than that from the magenta-flowered M. l. variegatus.
Iker Sanchez Marquez '26 and Caine Ryan '26. Iker (left) and Caine (right) worked together to replicate an experimental comparison of gene function, between two alleles of the anthocyanin-activating transcription factor gene MYB5a. They quantified anthocyanin by both digital image analysis and spectrophotometry. Both approaches confirmed the results of our initial experiment: Coding sequence from the yellow-flowered M. l. luteus, suprisingly, drives slightly but significantly more anthocyanin production than that from the magenta-flowered M. l. variegatus.
 Mussa Ghuza '25. Mussa was the greenhouse manager in spring 2024. He cared for a variety of monkeyflower and tobacco plants, and performed plant pollinations and seed collection.
Mussa Ghuza '25. Mussa was the greenhouse manager in spring 2024. He cared for a variety of monkeyflower and tobacco plants, and performed plant pollinations and seed collection.
 Nadiia Balbek '24. Nadiia helped to genetically map color patterning in hybrid monkeyflowers. To do this, she used Recombinant Inbred Lines (RILS) derived from a cross between the magenta-flowered M. l. variegatus and a yellow-flowered morph of M. cupreus. The F1 and F2 hybrids display complex color patterning, not seen in either parent. Repeated selfing of hundreds of F2s has generated a panel of RILs, some of which Nadiia crossed together to generate a diverse genetic mapping population which will focus on one or two of the patterning traits that differ between the starting RILs.
Nadiia Balbek '24. Nadiia helped to genetically map color patterning in hybrid monkeyflowers. To do this, she used Recombinant Inbred Lines (RILS) derived from a cross between the magenta-flowered M. l. variegatus and a yellow-flowered morph of M. cupreus. The F1 and F2 hybrids display complex color patterning, not seen in either parent. Repeated selfing of hundreds of F2s has generated a panel of RILs, some of which Nadiia crossed together to generate a diverse genetic mapping population which will focus on one or two of the patterning traits that differ between the starting RILs.
 Isa Lorenzetti '24. Isa advanced polyploidy research in Mimulus for the tetraploid M. cupreus and a hexaploid M. glabratus. She collected tissue for DNA sequencing, and extracted RNA from seven different tissue types for transcriptome sequencing. These collections are part of a larger effort by the monekyflower community to advance genomic resources in the genus, and will result in a high-quality genome for each species. Meanwhile, she took a first look at gene expression and copy number for two different genes in M. glabratus, MYB5a and GAPDH, as a case study of what happens to genes in hexaploids.
Isa Lorenzetti '24. Isa advanced polyploidy research in Mimulus for the tetraploid M. cupreus and a hexaploid M. glabratus. She collected tissue for DNA sequencing, and extracted RNA from seven different tissue types for transcriptome sequencing. These collections are part of a larger effort by the monekyflower community to advance genomic resources in the genus, and will result in a high-quality genome for each species. Meanwhile, she took a first look at gene expression and copy number for two different genes in M. glabratus, MYB5a and GAPDH, as a case study of what happens to genes in hexaploids.
 Abed Jomaa '25. Abed managed the research greenhouse for us in Summer 2023, including advancing our Recombinant Inbred Lines through selfing. He learned DNA and RNA extraction techniques, and will assisted with DNA extractions for genetic mapping later in the summer. He also contributed to our collaboration with the College of William and Mary to phenotype the collection of RILs.
Abed Jomaa '25. Abed managed the research greenhouse for us in Summer 2023, including advancing our Recombinant Inbred Lines through selfing. He learned DNA and RNA extraction techniques, and will assisted with DNA extractions for genetic mapping later in the summer. He also contributed to our collaboration with the College of William and Mary to phenotype the collection of RILs.
 Micaiah Newman '23. Micaiah extended a course-based research project from Plant Physiology, using a Q-Box photosynthesis system to measure rates of carbon assimilation and transpiration under varying levels of atmospheric CO2. Her study species was Eruca sativa (arugula). Despite the importance of understanding plant responses to elevated atmospheric carbon, in today's era of climate change, no publications to our knowledge have addressed this issue in Eruca sativa.
Micaiah Newman '23. Micaiah extended a course-based research project from Plant Physiology, using a Q-Box photosynthesis system to measure rates of carbon assimilation and transpiration under varying levels of atmospheric CO2. Her study species was Eruca sativa (arugula). Despite the importance of understanding plant responses to elevated atmospheric carbon, in today's era of climate change, no publications to our knowledge have addressed this issue in Eruca sativa.
 Kei Kim '22. Kei investigated the molecular mechanisms by which transcription factor MYB5a activates petal anthocyanin pigments in the magenta-flowered M. l. variegatus. She found additional evidence for what appears to be a post-transcriptional modification of the MYB5a mRNA transcript, and discovered that the functional divergence of MYB5a between M. l. variegatus and M. l. luteus is not due to changes in the coding sequence.
Kei Kim '22. Kei investigated the molecular mechanisms by which transcription factor MYB5a activates petal anthocyanin pigments in the magenta-flowered M. l. variegatus. She found additional evidence for what appears to be a post-transcriptional modification of the MYB5a mRNA transcript, and discovered that the functional divergence of MYB5a between M. l. variegatus and M. l. luteus is not due to changes in the coding sequence.
 Leah Samuels '22. Two models for how patterns develop are: (1) through a 'reaction-diffusion' system, or (2) through positional specification. We suspect that both types of pattern specification might be at work in determining the locations of spots seen in M. l. variegatus x M. cupreus hybrids, with the positional specification possibly being provided by the 'vascular bundles,' or veins, that are in the flower petals. To test this hypothesis, Leah developed protocols to chemically perturb vein development.
Leah Samuels '22. Two models for how patterns develop are: (1) through a 'reaction-diffusion' system, or (2) through positional specification. We suspect that both types of pattern specification might be at work in determining the locations of spots seen in M. l. variegatus x M. cupreus hybrids, with the positional specification possibly being provided by the 'vascular bundles,' or veins, that are in the flower petals. To test this hypothesis, Leah developed protocols to chemically perturb vein development.
 Ella Schochatovitz '22. Ella single-handedly designed and built our first ever CRISPR transgene, to knock out the anthocyanin-activating transcription factor MYB2b in M. cupreus. We eventually hope to obtain yellow-flowered M. cupreus that are as robust and easy to self-fertilize as our orange morph of M. cupreus. If yellow flowers are indeed produced, that would confirm our preliminary RNAi results suggesting that MYB2b is required for the orange petals normally found in this taxon.
Ella Schochatovitz '22. Ella single-handedly designed and built our first ever CRISPR transgene, to knock out the anthocyanin-activating transcription factor MYB2b in M. cupreus. We eventually hope to obtain yellow-flowered M. cupreus that are as robust and easy to self-fertilize as our orange morph of M. cupreus. If yellow flowers are indeed produced, that would confirm our preliminary RNAi results suggesting that MYB2b is required for the orange petals normally found in this taxon.
 Jackie Jones '22. Jackie performed numerous rounds of RNAi to target candidate pigmentation genes in both M. cupreus and M. naiandinus. Although she obtained several transgenic plants, none of them had an obviously disrupted pigmentation phenotype. The candidate genes are tandemly arrayed copies of MYB genes, and one possibility is that knocking down a single gene copy is insufficient to cause a phenotype because of functional redundancy across the copies. Another possibility is the known variability in strength of RNAi phenotypes, which might mean that we need a large number of RNAi lines on average to detect some lines with strong phenotypes... or, switch to CRISPR!
Jackie Jones '22. Jackie performed numerous rounds of RNAi to target candidate pigmentation genes in both M. cupreus and M. naiandinus. Although she obtained several transgenic plants, none of them had an obviously disrupted pigmentation phenotype. The candidate genes are tandemly arrayed copies of MYB genes, and one possibility is that knocking down a single gene copy is insufficient to cause a phenotype because of functional redundancy across the copies. Another possibility is the known variability in strength of RNAi phenotypes, which might mean that we need a large number of RNAi lines on average to detect some lines with strong phenotypes... or, switch to CRISPR!
Pandemic alums, summer 2020: Clockwise from top left: Joshua Shin '21 moved his microcopy setup to his rental house to continue his studies of petal patterns; Walker Orr '20 communicates with Prof. Cooley through the window; Ilona Wall '22 and Stephon Roberts '21 hold Zoom meetings to discuss their fruitfly research projects. After graduating in December 2020, Walker stayed on as a post-bacc lab tech through June, finally getting to work in the same room as real live labmates only during his final month. 
 Dan Thomas, Ph.D. Dan joined the lab in April 2018 and worked on a variety of genomic and transgenic projects. He completed a digital image analysis pipeline that allowed us to quantify spatially complex pigmentation patterns in a variegatus x cupreus mapping population, and contributed to molecular and transcriptomic analyses of pigmentation genes in several of the Mimulus taxa that we study.
Dan Thomas, Ph.D. Dan joined the lab in April 2018 and worked on a variety of genomic and transgenic projects. He completed a digital image analysis pipeline that allowed us to quantify spatially complex pigmentation patterns in a variegatus x cupreus mapping population, and contributed to molecular and transcriptomic analyses of pigmentation genes in several of the Mimulus taxa that we study.
 Andreas Guerrero '20. Andreas explored the effects of Myb3a on petal anthocyanin pigmentation in M. naiandinus. He used RNAi to knock down the gene in two different lines of M. naiandinus. He also started work on an overexpression construct, to drive expression of the naiandinus allele of Myb3a in species that normally lack petal anthocyanin pigmentation. Andreas is interested in bioinformatics and has been trying out some command-line work on our M. naiandinus transcriptome.
Andreas Guerrero '20. Andreas explored the effects of Myb3a on petal anthocyanin pigmentation in M. naiandinus. He used RNAi to knock down the gene in two different lines of M. naiandinus. He also started work on an overexpression construct, to drive expression of the naiandinus allele of Myb3a in species that normally lack petal anthocyanin pigmentation. Andreas is interested in bioinformatics and has been trying out some command-line work on our M. naiandinus transcriptome.
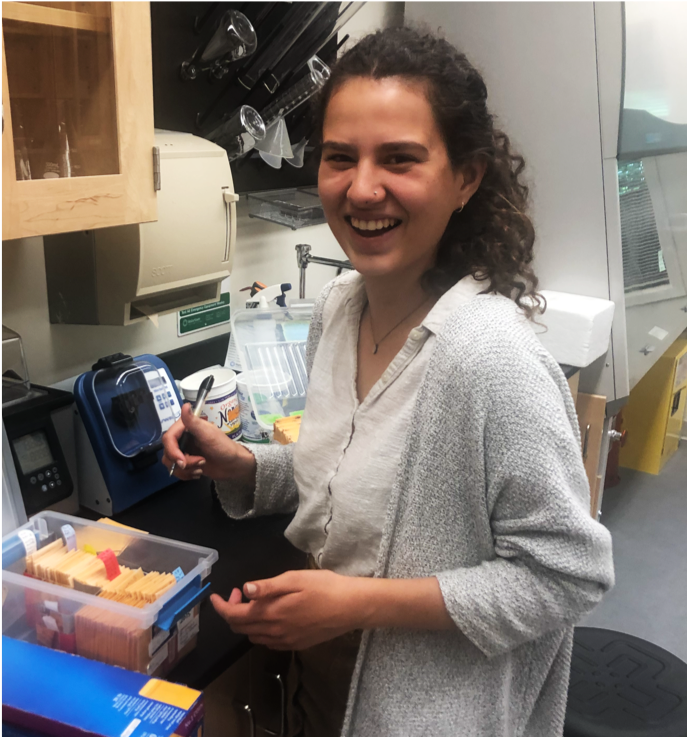 Bella Rivera '20. Bella was not a biology major - she completed an independently planned major in Global Health. But she also loves plants! She contributed to several of our research projects, primarily the effort to map complex patterning in variegatus x cupreus hybrids, and cared for our plants in the greenhouse for three years.
Bella Rivera '20. Bella was not a biology major - she completed an independently planned major in Global Health. But she also loves plants! She contributed to several of our research projects, primarily the effort to map complex patterning in variegatus x cupreus hybrids, and cared for our plants in the greenhouse for three years.
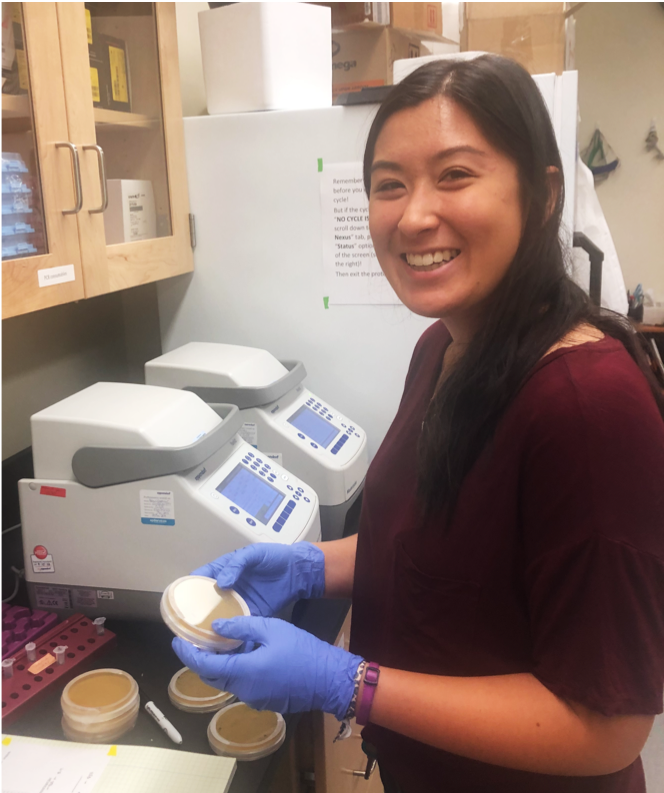 Ellen Hom '20. Ellen investigated two questions related to the gain of petal anthocyanin in M. l. variegatus: (1) Is overexpression of candidate gene Myb5 sufficient to activate petal anthocyanin in a Mimulus plant that normally lacks this pigmentation? (2) Why do we see what appear to be two distinct alleles of Myb5 in a highly inbred plant? Her results suggest that one of these "alleles" is present in the genome while the other may arise as a result of post-transcriptional editing.
Ellen Hom '20. Ellen investigated two questions related to the gain of petal anthocyanin in M. l. variegatus: (1) Is overexpression of candidate gene Myb5 sufficient to activate petal anthocyanin in a Mimulus plant that normally lacks this pigmentation? (2) Why do we see what appear to be two distinct alleles of Myb5 in a highly inbred plant? Her results suggest that one of these "alleles" is present in the genome while the other may arise as a result of post-transcriptional editing.
 Melia Matthews '20. Melia explored how complexity can arise from inter-genomic interactions, by genetically mapping the flower color patterning seen in the F1 and F2 hybrids of the two apparently solid-colored species M. l. variegatus and M. cupreus. In collaboration with Dan Thomas and Joshua Puzey's lab, she quantified petal phenotypes from an F2 mapping population, and used these in combination with Illumina sequence data to identify genomic regions responsible for elements of spatial pigment patterning.
Melia Matthews '20. Melia explored how complexity can arise from inter-genomic interactions, by genetically mapping the flower color patterning seen in the F1 and F2 hybrids of the two apparently solid-colored species M. l. variegatus and M. cupreus. In collaboration with Dan Thomas and Joshua Puzey's lab, she quantified petal phenotypes from an F2 mapping population, and used these in combination with Illumina sequence data to identify genomic regions responsible for elements of spatial pigment patterning.
 Maddy Boyle '20. In January-March 2019, Maddy constructed an RNAi transgene to knock down Myb3a in M. naiandinus, then handing off her project to Andreas to depart for a study abroad in Japan. She returned to the lab in fall 2019, to continue the investigation of RNAi knockdown effects on M. naiandinus.
Maddy Boyle '20. In January-March 2019, Maddy constructed an RNAi transgene to knock down Myb3a in M. naiandinus, then handing off her project to Andreas to depart for a study abroad in Japan. She returned to the lab in fall 2019, to continue the investigation of RNAi knockdown effects on M. naiandinus.
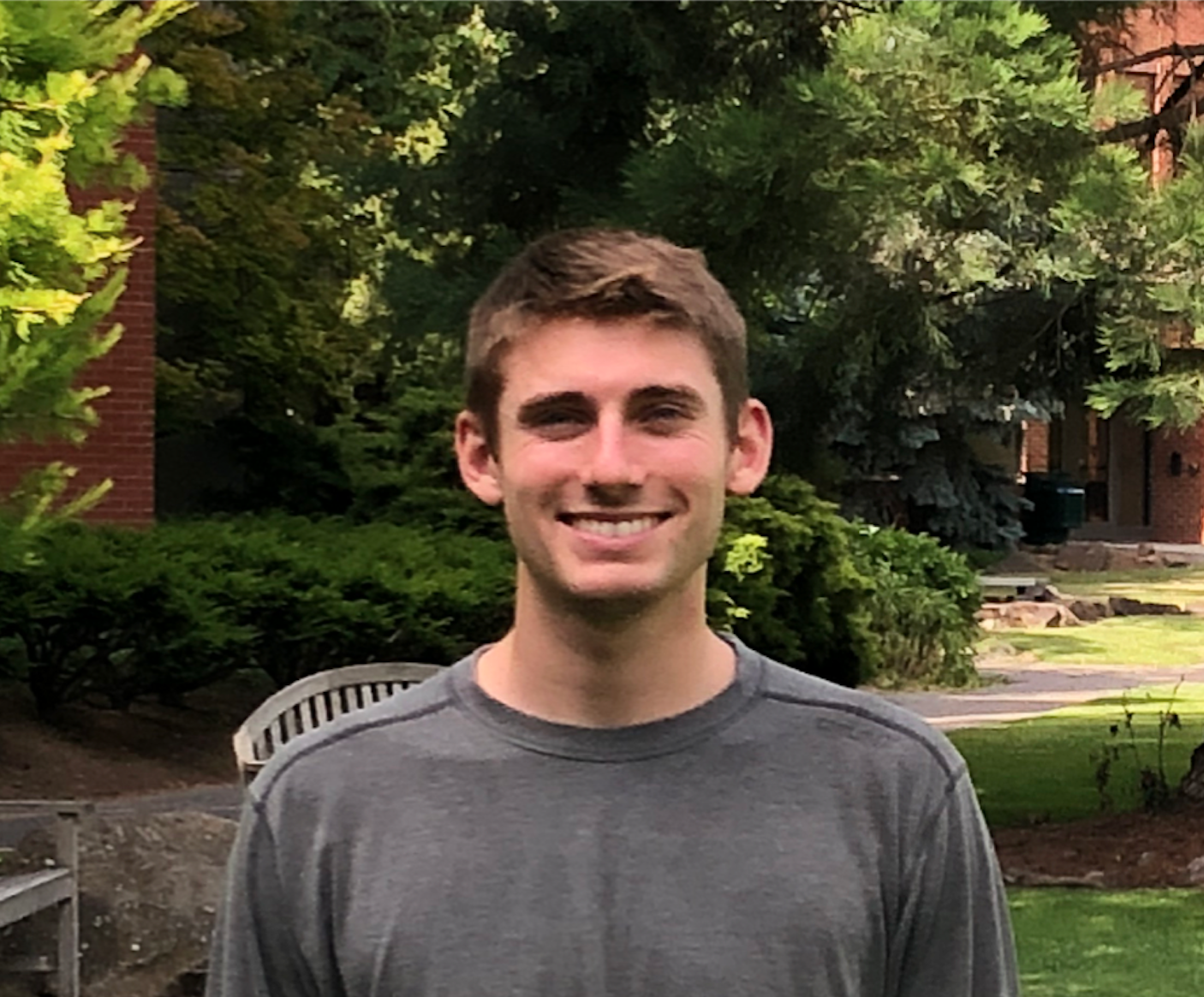 Jonah Rodewald '19. Jonah launched gene expression work in our least-studied species, M. naiandinus. In both M. naiandinus and M. cupreus, genomic region pla1 activates petal anthocyanins, but we didn't know whether the two species independently invented the idea, or whether a single ancestral mutation was shared between the species. Jonah found evidence for a partially shared genetic basis: M. cupreus appears to utilize both genes Myb2b and Myb3a, while M. naiandinus appears to rely solely on Myb3a.
Jonah Rodewald '19. Jonah launched gene expression work in our least-studied species, M. naiandinus. In both M. naiandinus and M. cupreus, genomic region pla1 activates petal anthocyanins, but we didn't know whether the two species independently invented the idea, or whether a single ancestral mutation was shared between the species. Jonah found evidence for a partially shared genetic basis: M. cupreus appears to utilize both genes Myb2b and Myb3a, while M. naiandinus appears to rely solely on Myb3a.
 Ashley Person '19. Ashley used RNAi to knock down candidate petal anthocyanin genes Myb2b and Myb3a in the orange-flowered species M. cupreus. She documented a range of reduced-pigment RNAi phenotypes. These phenotypes were transient within a plant, possibly due to epigenetic silencing. Ashley also contributed to building an overexpression transgene to determine whether the variegatus allele of Myb5 is sufficient to activate petal anthocyanin.
Ashley Person '19. Ashley used RNAi to knock down candidate petal anthocyanin genes Myb2b and Myb3a in the orange-flowered species M. cupreus. She documented a range of reduced-pigment RNAi phenotypes. These phenotypes were transient within a plant, possibly due to epigenetic silencing. Ashley also contributed to building an overexpression transgene to determine whether the variegatus allele of Myb5 is sufficient to activate petal anthocyanin.
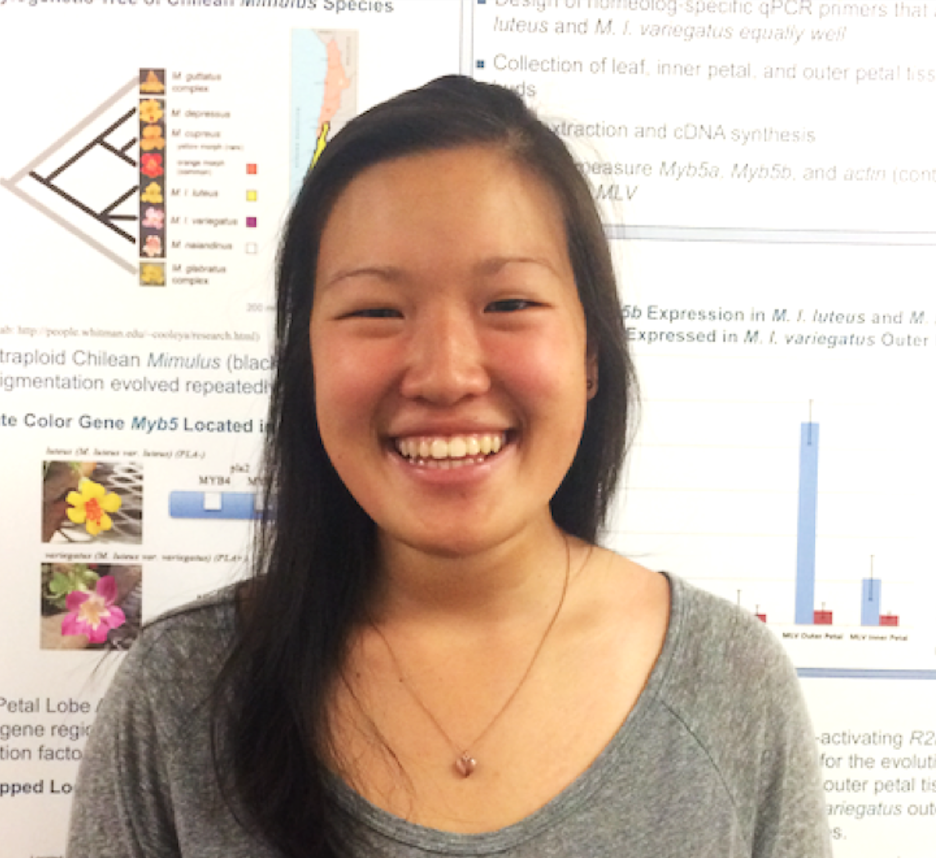 Johanna Au '19. Johanna helped build RNAi constructs to silence two genes, Myb2b and and Myb3a, that are our top candidates for causing the evolutionary gain of petal anthocyanin pigmentation in the orange-flowered Mimulus cupreus. She used a Gateway strategy, in which fragments of each gene are directionally cloned into the pENTR entry vector and then recombined in to a destination vector that will be used for transformation.
Johanna Au '19. Johanna helped build RNAi constructs to silence two genes, Myb2b and and Myb3a, that are our top candidates for causing the evolutionary gain of petal anthocyanin pigmentation in the orange-flowered Mimulus cupreus. She used a Gateway strategy, in which fragments of each gene are directionally cloned into the pENTR entry vector and then recombined in to a destination vector that will be used for transformation.
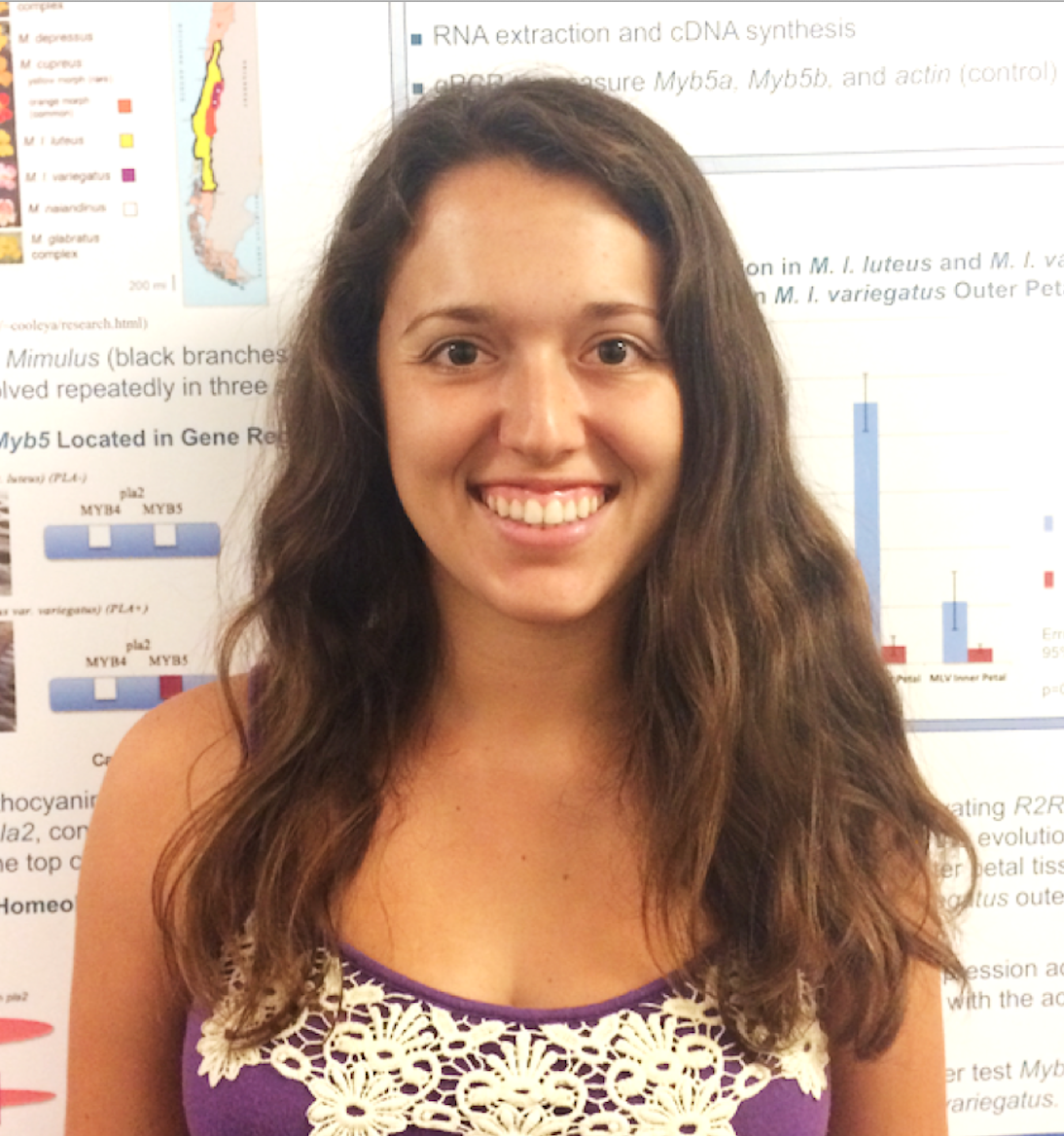 Jessie Friedman '18. A recently-constructed Mimulus cupreus transcriptome provided coding sequence for petal pigment candidate genes Myb2b and Myb3a. To fully evaluate the functions of these genes, however, we need to know their upstream regulatory sequences so that we can build transgenic constructs driven by the native promoter of each gene. Jessie leveraged existing genomic resources to design primers to "fish out" promoter sequence from M. cupreus.
Jessie Friedman '18. A recently-constructed Mimulus cupreus transcriptome provided coding sequence for petal pigment candidate genes Myb2b and Myb3a. To fully evaluate the functions of these genes, however, we need to know their upstream regulatory sequences so that we can build transgenic constructs driven by the native promoter of each gene. Jessie leveraged existing genomic resources to design primers to "fish out" promoter sequence from M. cupreus.
 Kuenzang Om '18. Kuenzang used transgenics to functionally test the role of candidate gene Myb5 in petal lobe anthocyanin (PLA) pigmentation. In this photo she is transforming the purple-flowered M. l. variegatus with an RNAi construct to silence Myb5. She found that the resulting transgenic offspring had white flowers, indicating that Myb5 is indeed necessary for the recently-evolved trait of PLA in M. l. variegatus.
Kuenzang Om '18. Kuenzang used transgenics to functionally test the role of candidate gene Myb5 in petal lobe anthocyanin (PLA) pigmentation. In this photo she is transforming the purple-flowered M. l. variegatus with an RNAi construct to silence Myb5. She found that the resulting transgenic offspring had white flowers, indicating that Myb5 is indeed necessary for the recently-evolved trait of PLA in M. l. variegatus.
 Erin Minus '18. Erin continued the work of Aaron Williams '17, using digital image analysis in ArcGIS to quantify spatially complex pigment patterns in an F2 hybrid population derived from an M. cupreus x M. l. variegatus cross. You can see the ArcGIS program running in the background of this photo. Erin will compare phenotypes she obtains to the genotypes from our first sequenced individuals (96 F2 plants) in order to genetically map some of the traits.
Erin Minus '18. Erin continued the work of Aaron Williams '17, using digital image analysis in ArcGIS to quantify spatially complex pigment patterns in an F2 hybrid population derived from an M. cupreus x M. l. variegatus cross. You can see the ArcGIS program running in the background of this photo. Erin will compare phenotypes she obtains to the genotypes from our first sequenced individuals (96 F2 plants) in order to genetically map some of the traits.
 Taylor Wilke '18. Taylor used the same F2 population as Erin to investigate a different trait, yellow carotenoid pigment. When Mimulus cupreus gained petal anthocyanin, it kept its carotenoids; the combination of the two pigments are what creates its orange color. Mimulus l. variegatus, in contrast, gained anthocyanin but lost floral carotenoids. Carotenoid abundance segregates in our F2 mapping population. Suprisingly little is known about the genetics of carotenoid regulation, and we hope to discover some of the genes involved over the course of this mapping project.
Taylor Wilke '18. Taylor used the same F2 population as Erin to investigate a different trait, yellow carotenoid pigment. When Mimulus cupreus gained petal anthocyanin, it kept its carotenoids; the combination of the two pigments are what creates its orange color. Mimulus l. variegatus, in contrast, gained anthocyanin but lost floral carotenoids. Carotenoid abundance segregates in our F2 mapping population. Suprisingly little is known about the genetics of carotenoid regulation, and we hope to discover some of the genes involved over the course of this mapping project.
 Rachel Eguia '17. Rachel worked on building transgenes to test the function of candidate pigmentation gene Myb5. The eventual goal is to silence Myb5 in plants that have petal anthocyanin pigmentation, and activate Myb5 in plants that lack petal anthocyanin pigmentation, in order to test the hypothesis that Myb5 is both necessary and sufficient for the recent gain of petal anthocyanins in the purple-flowered Mimulus luteus var. variegatus.
Rachel Eguia '17. Rachel worked on building transgenes to test the function of candidate pigmentation gene Myb5. The eventual goal is to silence Myb5 in plants that have petal anthocyanin pigmentation, and activate Myb5 in plants that lack petal anthocyanin pigmentation, in order to test the hypothesis that Myb5 is both necessary and sufficient for the recent gain of petal anthocyanins in the purple-flowered Mimulus luteus var. variegatus.
 Aaron Williams '17. Crossing a plant with solid purple flowers (M. l. variegatus) to a plant with solid orange flowers (M. cupreus) yields a hybrid with complex patterns of red blotches overlaid on a cream background. Little is known about the developmental regulation of "speckliness," and it is difficult to measure using traditional means. Aaron helped develop a digital image analysis approach for extracting quantitative trait values from the complex spatial variation seen in an F2 hybrid population.
Aaron Williams '17. Crossing a plant with solid purple flowers (M. l. variegatus) to a plant with solid orange flowers (M. cupreus) yields a hybrid with complex patterns of red blotches overlaid on a cream background. Little is known about the developmental regulation of "speckliness," and it is difficult to measure using traditional means. Aaron helped develop a digital image analysis approach for extracting quantitative trait values from the complex spatial variation seen in an F2 hybrid population.
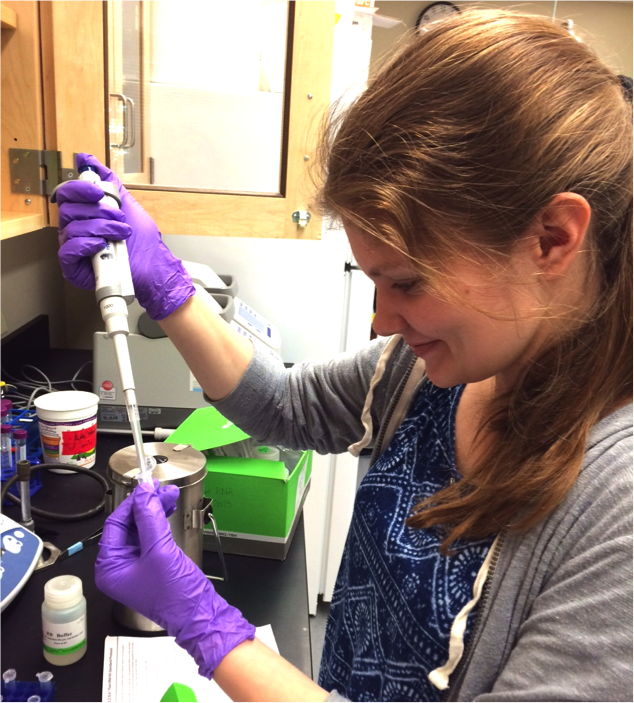 Anne Vonada '17. The recent gain of petal anthocyanins in the orange-flowered M. cupreus is controlled by a single locus that we call pla1. This region of the genome is unlinked to Myb5 (the top candidate in M. l. variegatus), but it does contain numerous Myb transcription factors that are similarly good candidates for activating petal pigmentation in M. cupreus. Anne Vonada used genomics, transcriptomics, and RT-PCR to evaluate candidate genes at pla1, and was able to narrow down the list from six genes to two: Myb2b and Myb3a.
Anne Vonada '17. The recent gain of petal anthocyanins in the orange-flowered M. cupreus is controlled by a single locus that we call pla1. This region of the genome is unlinked to Myb5 (the top candidate in M. l. variegatus), but it does contain numerous Myb transcription factors that are similarly good candidates for activating petal pigmentation in M. cupreus. Anne Vonada used genomics, transcriptomics, and RT-PCR to evaluate candidate genes at pla1, and was able to narrow down the list from six genes to two: Myb2b and Myb3a.
 Jessica Palacios '16. Previous work in the luteus complex of Mimulus has utilized a single inbred line as the representative of the species. But just how representative is that line? Jessica launched an exploration of the wider range of diversity within several species, which revealed consistent trends as well as important differences between our inbred lines and a panel of wild-collected accessions.
Jessica Palacios '16. Previous work in the luteus complex of Mimulus has utilized a single inbred line as the representative of the species. But just how representative is that line? Jessica launched an exploration of the wider range of diversity within several species, which revealed consistent trends as well as important differences between our inbred lines and a panel of wild-collected accessions.
 Jeremy Nolan '16. Jeremy is interested in the physiological consequences of environmental pollution. He modified a plant incubator to serve as a CO2 chamber, in order to test the effects of elevated CO2 on plant growth and photosynthesis across a diverse collection of Mimulus accessions from Chile.
Jeremy Nolan '16. Jeremy is interested in the physiological consequences of environmental pollution. He modified a plant incubator to serve as a CO2 chamber, in order to test the effects of elevated CO2 on plant growth and photosynthesis across a diverse collection of Mimulus accessions from Chile.
 Allison Eggert '16. The luteus group of Mimulus is tetraploid, and contains two "homeologous" (duplicate) copies of the Myb5 gene. Myb5a is tightly linked to the gain of petal pigmentation in M. l. variegatus, while Myb5b is unlinked. Surprisingly, they are 100% identical from start codon through 3'UTR. Divergence in the 5'UTR allowed Allison to design homeolog-specific primers. She found that Myb5a has high expression in anthocyanin-pigmented petals, while Myb5b has uniformly low expression across tissues and species.
Allison Eggert '16. The luteus group of Mimulus is tetraploid, and contains two "homeologous" (duplicate) copies of the Myb5 gene. Myb5a is tightly linked to the gain of petal pigmentation in M. l. variegatus, while Myb5b is unlinked. Surprisingly, they are 100% identical from start codon through 3'UTR. Divergence in the 5'UTR allowed Allison to design homeolog-specific primers. She found that Myb5a has high expression in anthocyanin-pigmented petals, while Myb5b has uniformly low expression across tissues and species.
 Kimberly Stanton. Kimmy, a Reed College ('14) graduate, worked as a technician in the Cooley lab before going on to graduate school in evolutionary biology at Duke University. She developed a transcriptome-supported approach to identifying qPCR reference genes for Mimulus, and worked towards transgenic transformation in M. cupreus. Here, Kimmy prepares plants for a vacuum infiltration to drive Agrobacteria into the floral bud tissue, as part of plant transformation. (Photo Credit: Larry North)
Kimberly Stanton. Kimmy, a Reed College ('14) graduate, worked as a technician in the Cooley lab before going on to graduate school in evolutionary biology at Duke University. She developed a transcriptome-supported approach to identifying qPCR reference genes for Mimulus, and worked towards transgenic transformation in M. cupreus. Here, Kimmy prepares plants for a vacuum infiltration to drive Agrobacteria into the floral bud tissue, as part of plant transformation. (Photo Credit: Larry North)
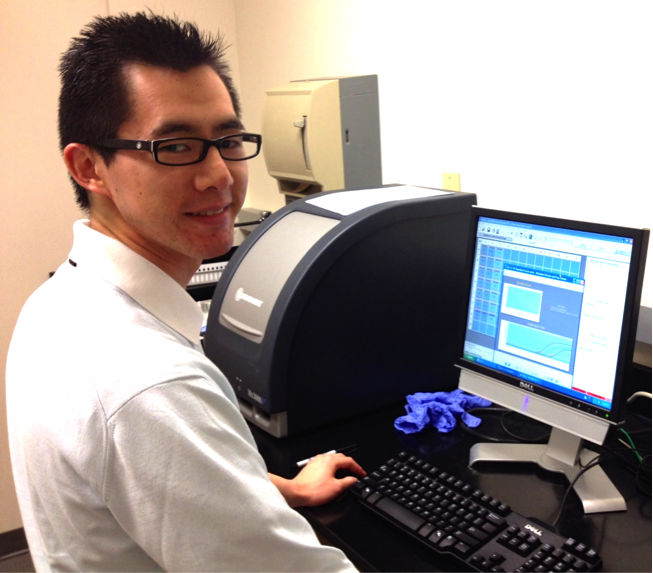 Philip Cheng '15. Philip used quantitative RT-PCR to measure expression patterns of candidate pigmentation genes Myb4 and Myb5 in M. l. luteus and M. l. variegatus. Here he is setting up a run on the qPCR machine. Philip discovered that Myb4 has almost no expression in leaf, petal, or nectar guide tissue; in contrast, Myb5 expression is high in anthocyanin-pigmented tissues. This strongly supports Myb5 as the causal gene for pigment gain in M. l. variegatus.
Philip Cheng '15. Philip used quantitative RT-PCR to measure expression patterns of candidate pigmentation genes Myb4 and Myb5 in M. l. luteus and M. l. variegatus. Here he is setting up a run on the qPCR machine. Philip discovered that Myb4 has almost no expression in leaf, petal, or nectar guide tissue; in contrast, Myb5 expression is high in anthocyanin-pigmented tissues. This strongly supports Myb5 as the causal gene for pigment gain in M. l. variegatus.
 Marijke Wijnen '15. Celine's research revealed an anthocyanin-producing stress response following an (unplanned!) heat wave that imposed both heat and drought. Marijke followed up on this work, using controlled treatments to disentangle the effects of these two environmental stressors. She found that drought, but not heat, induced vegetative anthocyanin pigments. Interestingly, the heat treatment resulted in an increased proportion of deformed flowers - possibly a Heat Shock Protein effect..? Here she measures photosynthesis in the greenhouse.
Marijke Wijnen '15. Celine's research revealed an anthocyanin-producing stress response following an (unplanned!) heat wave that imposed both heat and drought. Marijke followed up on this work, using controlled treatments to disentangle the effects of these two environmental stressors. She found that drought, but not heat, induced vegetative anthocyanin pigments. Interestingly, the heat treatment resulted in an increased proportion of deformed flowers - possibly a Heat Shock Protein effect..? Here she measures photosynthesis in the greenhouse.
 Celine Valentin '14. Celine found that the yellow-flowered M. l. luteus and the orange-flowered, more anthocyanin-rich M. cupreus responded differently to an intense week-long heat wave: the former produced significantly more vegetative anthocyanin pigments. Such environmentally-induced vegetative anthocyanins are considered a sign of plant stress. M. l. luteus therefore appears to have been more stressed during the heat wave. We hypothesize that M. cupreus, in contrast, may be "pre-protected" by its higher constitutive levels of anthocyanin pigments.
Celine Valentin '14. Celine found that the yellow-flowered M. l. luteus and the orange-flowered, more anthocyanin-rich M. cupreus responded differently to an intense week-long heat wave: the former produced significantly more vegetative anthocyanin pigments. Such environmentally-induced vegetative anthocyanins are considered a sign of plant stress. M. l. luteus therefore appears to have been more stressed during the heat wave. We hypothesize that M. cupreus, in contrast, may be "pre-protected" by its higher constitutive levels of anthocyanin pigments.
 Leah Mohtes-Chan '14 (left) and Janae Edelson '14 (right). Leah and Janae launched the molecular lab - Step 1, unpack a LOT of boxes! They extracted DNA from M. l. luteus and prepared it for genomic sequencing at Duke University, which led to the sequencing of the first ever Chilean Mimulus genome. They also used semi-quantitative RT-PCR to discover that two tandemly duplicated candidate pigmentation genes, Myb4 and Myb5, have diverged in function: Myb5 is expressed in both petal and leaf tissue, while Myb4 is not.
Leah Mohtes-Chan '14 (left) and Janae Edelson '14 (right). Leah and Janae launched the molecular lab - Step 1, unpack a LOT of boxes! They extracted DNA from M. l. luteus and prepared it for genomic sequencing at Duke University, which led to the sequencing of the first ever Chilean Mimulus genome. They also used semi-quantitative RT-PCR to discover that two tandemly duplicated candidate pigmentation genes, Myb4 and Myb5, have diverged in function: Myb5 is expressed in both petal and leaf tissue, while Myb4 is not.
 Sage Stutsman '13. Founding member of the Cooley lab, Sage planted and tended the very first monkeyflower population at Whitman. She is especially interested in hydroponics and developed a hydroponics system to study the effects of different nutrient treatments on plant growth and development.
Sage Stutsman '13. Founding member of the Cooley lab, Sage planted and tended the very first monkeyflower population at Whitman. She is especially interested in hydroponics and developed a hydroponics system to study the effects of different nutrient treatments on plant growth and development.